Arabidopsis contains hundreds of ribosomal DNA copies organized within the nucleolar organizing regions (NORs) in chromosomes 2 and 4. There are four major types of variants of rDNA, VAR1–4, based on the polymorphisms of 3’ external transcribed sequences. The variants are known to be differentially expressed during plant development.
We created a mutant by the CRISPR-Cas9-mediated excision of ~ 25 nt from predominantly NOR4 ribosomal DNA copies, obtaining mosaic mutational events on ~ 5% of all rDNA copies. The excised region consists of P-loop and Helix-82 segments of 25S rRNA.
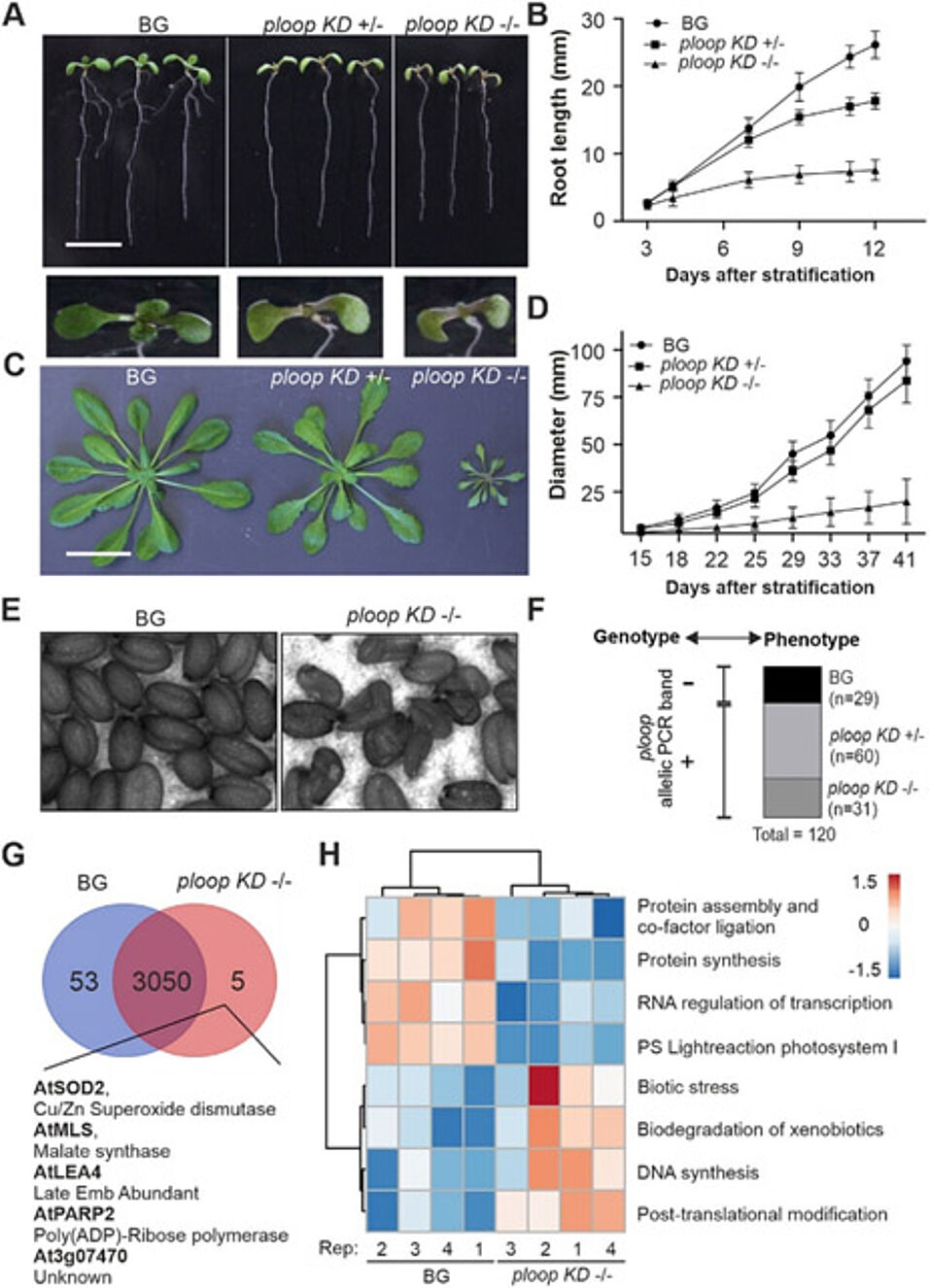
The mutation led to allelic, dosage-dependent defects marked by lateral root inhibition, reduced size, and pointy leaves, all previously observed for defective ribosomal function. The mutation in NOR4 led to dosage compensation from the NOR2 copies by elevated expression of VAR1 in mutants and further associated single-nucleotide variants, thus, resulting in altered rRNA sub-population. Furthermore, the mutants exhibited rRNA maturation defects specifically in the minor pathway typified by 32S pre-rRNA accumulation.
Density-gradient fractionation and subsequent RT-PCR of rRNA analyses revealed that mutated copies were not incorporated into the translating ribosomes. The mutants in addition displayed an elevated autophagic flux as shown by the autophagic marker GFP-ATG8e, likely related to ribophagy.
Read the full text here: