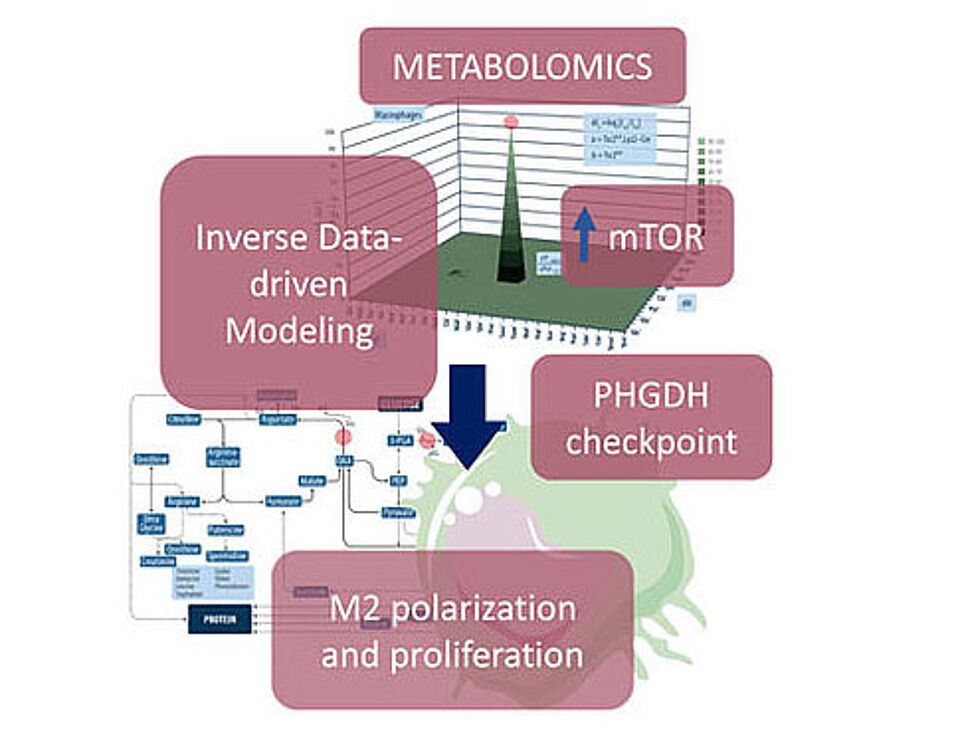
Read full paper for free on www.cell.com
Press coverage:
- Die Entwicklung von Krebszellen verhindern on univie.ac.at
- Die Entwicklung von Krebszellen verhindern on science.apa.at
- Forscher verhindern Unterstützung von Tumoren durch Riesenfresszellen on science.apa.at
- Fresszellen am Fördern von Krebs hindern on diepresse.com
- Biochemischer Schalter gegen Krebs on chemiextra.com
- Die Entwicklung von Krebszellen verhindern on analytik.de
- Targeting the cancer microenvironment on bioportfolio.com
- Targeting the cancer microenvironment on sciencedaily.com
- Targeting the cancer microenvironment on worldpharmanews.com