Computational methods for systems biology and biotechnology - Steffen Waldherr group
Room: 3.044
Djerassiplatz 1
1030 Vienna, Austria
T: +43-1-4277-76580
steffen.waldherr@univie.ac.at
Research topics
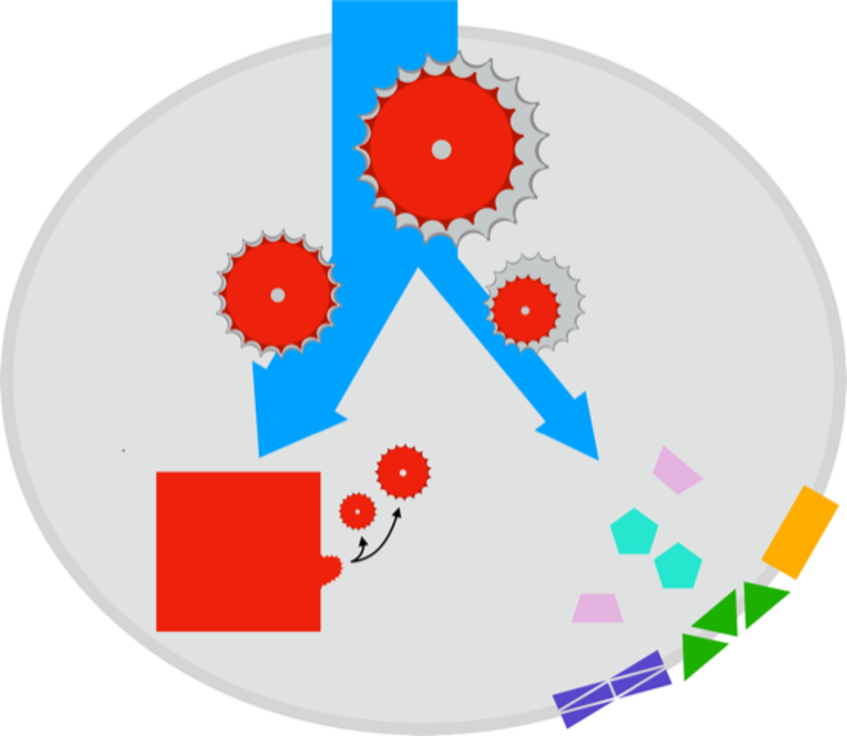
Dynamic flux balance and resource allocation models
Dynamic flux balance and resource allocation models
Dynamic flux balance analysis and resource allocation models are computational models used in systems biology and biotechnology to describe the metabolism and enzyme expression of cells. These models use optimization approaches to determine the optimal metabolic flux distribution and allocation of resources, such as nutrients or energy, to different cellular processes.
Our research in dynamic flux balance and resource allocation models is focused on developing more accurate and efficient computational methods, as well as constructing and applying these models to study a wide range of biological systems, including biotechnologcial processes, microbial communities, and plants. We also study the integration of multi-omics data to better predict cellular metabolism and physiology under different conditions.
Selected publications:
- De Becker, K., Totis, N., Bernaerts, K., & Waldherr, S. (2022). Using resource constraints derived from genomic and proteomic data in metabolic network models. Current Opinion in Systems Biology, 29, 100400. doi.org/10.1016/j.coisb.2021.100400
- Waldherr, S., Oyarzún, D. A., and Bockmayr, A. (2015). Dynamic optimization of metabolic networks coupled with gene expression. Journal of Theoretical Biology 365:469–485.
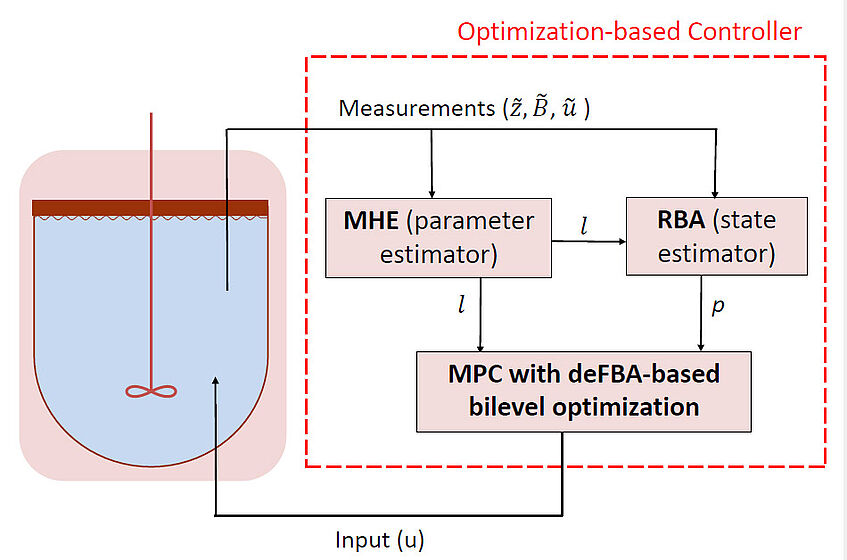
Bioprocess optimization and control with constraint-based models
Bioprocess optimization and control with constraint-based models
Constraint-based models, such as flux balance analysis and its extensions, have emerged as powerful tools for bioprocess optimization. In our research, we are using dynamic flux balance analysis and its extensions to maximize the yield or productivity of a desired product, such as biofuels, pharmaceuticals, or chemicals. Such a maximization can be achieved by optimization of process condition or by dynamic feedback control.
Selected publications:
Jabarivelisdeh, B., Carius, L., Findeisen, R., & Waldherr, S. (2020). Adaptive predictive control of bioprocesses with constraint-based modeling and estimation. Computers & Chemical Engineering, 135, 106744. doi.org/10.1016/j.compchemeng.2020.106744
Jabarivelisdeh, B., & Waldherr, S. (2018). Optimization of bioprocess productivity based on metabolic-genetic network models with bilevel dynamic programming. Biotechnology and Bioengineering, 115(7), 1829–1841. doi.org/10.1002/bit.26599
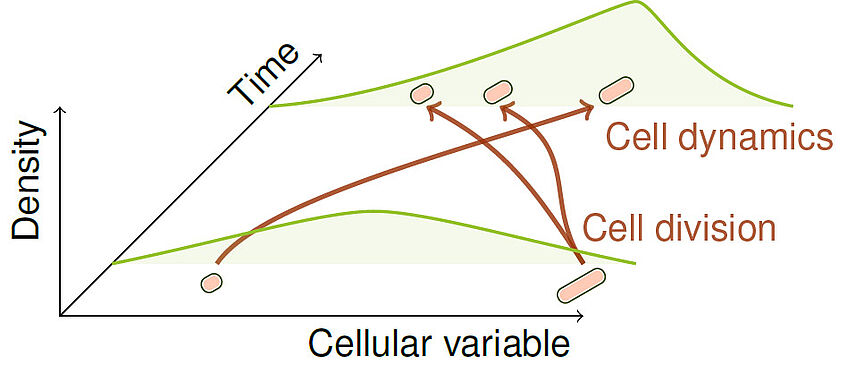
Modelling and estimation of heterogeneous cell populations
Modelling and estimation of heterogeneous cell populations
We develop individual-based and population balance models of heterogeneous cell populations, where cells differ in their characteristics, such as gene expression, protein activity, or metabolic state, which can impact physiology and response to extracellular stimuli on the cell population level. We also develop state estimators and parameter identification algorithms for such models that rely on so-called population snapshot data, which are high-throughput measurements of a reduced number of variables per cell, with different population samples at different time-points.
Selected publications:
- Küper, A., Dürr, R., and Waldherr, S. (2019): "Dynamic Density Estimation in Heterogeneous Cell Populations". IEEE Control Systems Letters 3:242–247.
- Waldherr, S. (2018). Estimation methods for heterogeneous cell population models in systems biology. Journal of the Royal Society Interface 15:20180530.
- Imig, D., Pollak, N., Strecker, T., Scheurich, P., Allgöwer, F., and Waldherr, S. (2015). An individual-based simulation framework for dynamic, heterogeneous cell populations during extrinsic stimulations. J. Coupled Syst. Multiscale Dyn. 3:122–134.
Machine learning for biochemical data and processes
Machine learning for biochemical data and processes
We are applying various machine learning approaches, such as Gaussian processes, neural networks, or hybrid models to the analysis of biological and biochemical data, including metabolomics and proteomics. We are particularly interested in time series data and models that allow to link data to biological insight. The main objective is to use such models to improve understanding and prediction of biological processes and dynamics.
Selected publications:
Sidak, D., Schwarzerová, J., Weckwerth, W., & Waldherr, S. (2022). Interpretable machine learning methods for predictions in systems biology from omics data. Frontiers in Molecular Biosciences, 9, 926623. doi.org/10.3389/fmolb.2022.926623
Di Caprio, U., Wu, M., Vermeire, F., Van Gerven, T., Hellinckx, P., Waldherr, S., Kayahan, E., & Leblebici, M. E. (2023). Predicting overall mass transfer coefficients of CO2 capture into monoethanolamine in spray columns with hybrid machine learning. Journal of CO2 Utilization, 70, 102452. doi.org/10.1016/j.jcou.2023.102452
