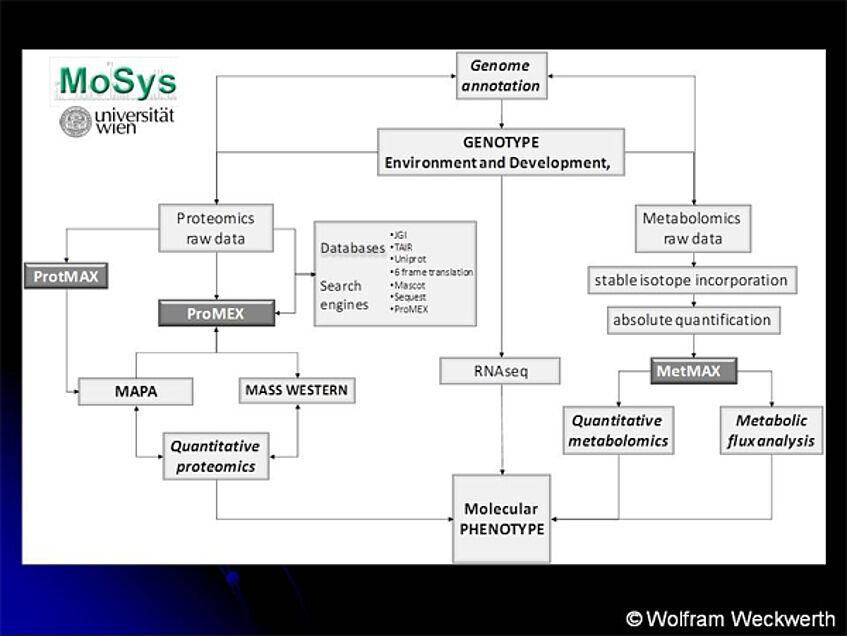
Strategy for functional interpretation of genotype data and environmental phenotypes based on molecular data
Strategy for functional interpretation of genotype data and environmental phenotypes based on molecular data
- for details see: Targeted proteomics for Chlamydomonas reinhardtii combined with rapid subcellular protein fractionation, metabolomics and metabolic flux analyses. Wienkoop S, Weiss J, May P, Kempa S, Irgang S, Recuenco-Munoz L, Pietzke M, Schwemmer T, Rupprecht J, Egelhofer V, Weckwerth W. Mol Biosyst. 2010 Jun 18;6(6):1018-31.
... Abstract
Profiling Platform MoSys to combine proteomics-assisted genome annotation (May et al., 2008), targeted proteomics (Mass Western (Wienkoop and Weckwerth, 2006; Lehmann et al., 2008; Wienkoop et al., 2008a)), subcellular fractionation, untargeted proteomics (MAPA: mass accuracy precursor alignment (Hoehenwarter et al., 2008)) for analysis of dynamic proteomes as well as to integrate the data with metabolomics and metabolic flux data (Weckwerth, 2008). The following algorithms were used to assemble the complete data set presented in this study: PROTMAX for the MAPA approach (Hoehenwarter et al., 2008), METMAX for the metabolomics and mass isotopomer analysis (Kempa et al., 2009) and PROMEX (http://www.promexdb.org) for proteomics data assembly, storage and spectral comparison (Hummel et al., 2007).