Unpredictability of Metabolism – a genotype-phenotype equation
Unpredictability of Metabolism – a genotype-phenotype equation
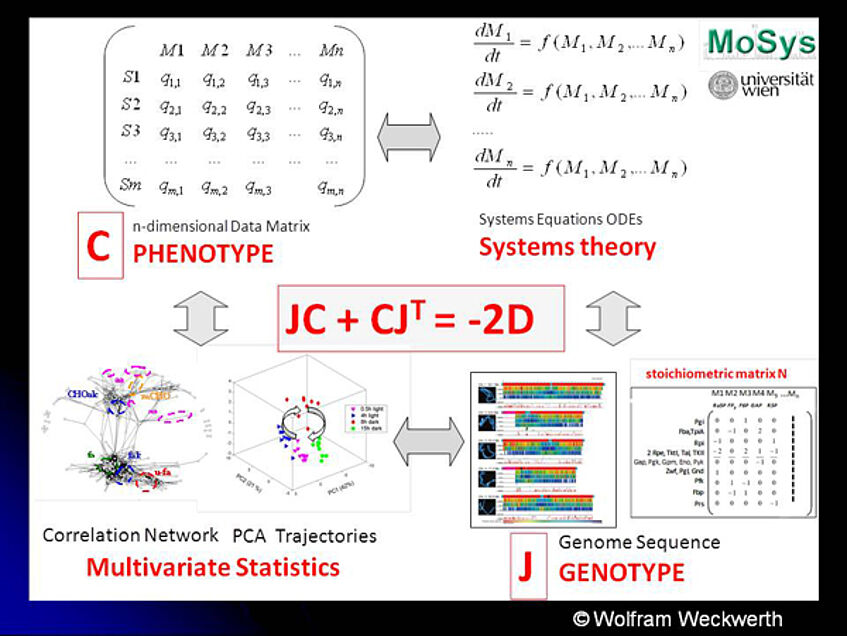
- for details see: Unpredictability of metabolism-the key role of metabolomics science in combination with next-generation genome sequencing. Weckwerth W. Anal Bioanal Chem. 2011 Jun;400(7):1967-78. ... Abstract Pubmed
Next generation sequencing (NGS) provides technologies to sequence whole prokaryotic and eukaryotic genomes in days, to perform genome-wide association (GWA) studies, chromatin immunoprecipitation followed by sequencing (ChIP-seq) and RNA sequencing (RNA-seq) for transcriptome studies. An exponential growing volume of sequence data can be anticipated, yet, functional interpretation does not keep pace with the amount of data produced. In principle, these data contain all the secrets of living systems, the genotype-phenotype-relationship. Firstly, it is possible to derive the structure and connectivity of the metabolic network from the genotype of an organism in form of the stoichiometric matrix N. This is, however, static information. Strategies for modelling and prediction of dynamic metabolic networks based on genome sequences need to be applied. Despite very few examples it becomes obvious how difficult the functional interpretation of newly sequenced genomes and the prediction of the corresponding dynamic metabolic networks are. Consequently, metabolomics science – the quantitative measurement of metabolism in conjunction with metabolic modelling - is a key discipline for the functional interpretation of whole genomes and especially for testing the numerical predictions of metabolism based on genome-scale metabolic network models. In this context, a systematic equation is derived based on metabolomics covariance data and the genome-scale stoichiometric matrix which describes the genotype-phenotype relationship.